At Qure, we regularly work on segmentation and object detection problems and we were therefore interested in reviewing the current state of the art.
In this post, I review the literature on semantic segmentation. Most research on semantic segmentation use natural/real world image datasets. Although the results are not directly applicable to medical images, I review these papers because research on the natural images is much more mature than that of medical images.
Post is oraganized as follows: I first explain the semantic segmentation problem, give an overview of the approaches and summarize a few interesting papers.
In a later post, I’ll explain why medical images are different from natural images and examine how the approaches from this review fare on a dataset representative of medical images.
What exactly is semantic segmentation?
Semantic segmentation is understanding an image at pixel level i.e, we want to asign each pixel in the image an object class. For example, check out the following images.

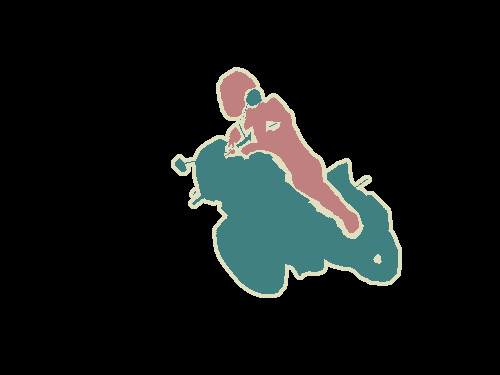
Left
: Input image.
Right
: It's semantic segmentation. Source.
Apart from recognising the bike and the person riding it, we also have to delineate the boundaries of each object. Therefore, unlike classification, we need dense pixel-wise predictions from our models.
VOC2012 and MSCOCO are the most important datasets for semantic segmentation.
What are the different approaches?
Before deep learning took over computer vision, people used approaches like TextonForest and Random Forest based classifiers for semantic segmentation. As with image classification, convolutional neural networks (CNN) have had enormous success on segmentation problems.
One of the popular initial deep learning approaches was patch classification where each pixel was separately classified into classes using a patch of image around it. Main reason to use patches was that classification networks usually have full connected layers and therefore required fixed size images.

















